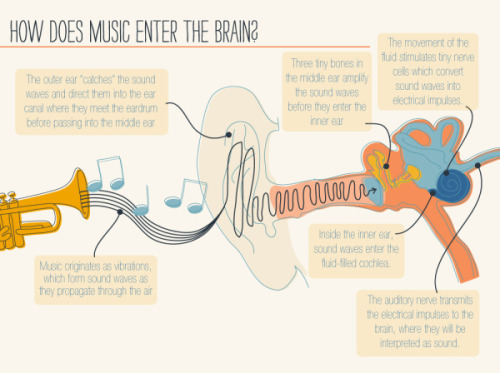
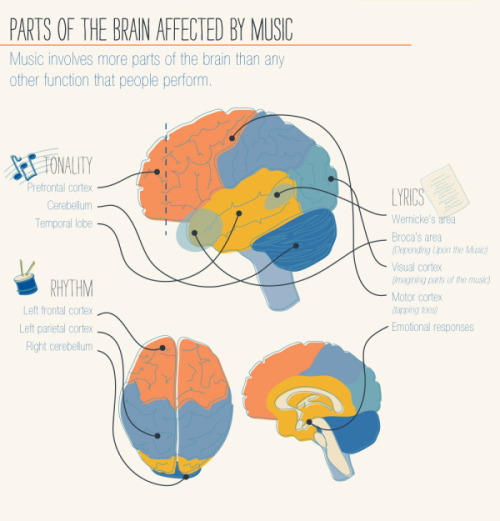
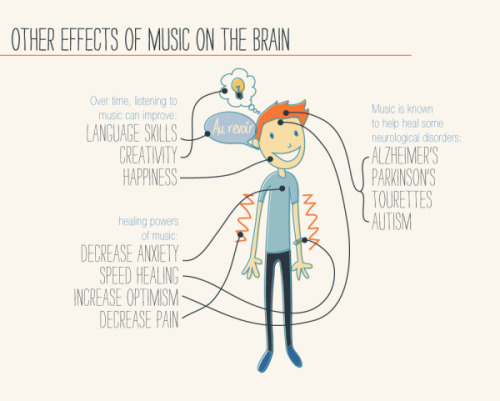
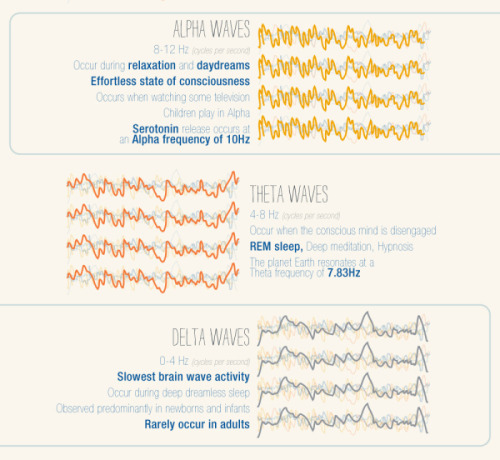
It’s A Textbook Moment Centuries In The Making: More Than 200 Years After Scientists Started Investigating

It’s a textbook moment centuries in the making: more than 200 years after scientists started investigating how water molecules conduct electricity, a team has finally witnessed it happening first-hand.
It’s no surprise that most naturally ocurring water conducts electricity incredibly well - that’s a fact most of us have been taught since primary school. But despite how fundamental the process is, no one had been able to figure out how it actually happens on the atomic level.
“This fundamental process in chemistry and biology has eluded a firm explanation,” said one of the team, Anne McCoy from the University of Washington. “And now we have the missing piece that gives us the bigger picture: how protons essentially ‘move’ through water.”
Continue Reading.
More Posts from Contradictiontonature and Others
Dive deep into Episode 05 of #ShelfLife to discover the various technologies that have helped humans map the sky around us for eons.

From sundials to mega-powered modern telescopes, tools for stargazing allow us to understand the universe—and our place within it. Season 2 begins on November 1.


Even after someone is declared dead, life continues in the body, suggests a surprising new study with important implications.
Gene expression — when information stored in DNA is converted into instructions for making proteinsor other molecules — actually increases in some cases after death, according to the new paper, which tracked postmortem activity and is published in the journal Open Biology.
“Not all cells are ‘dead’ when an organism dies,” senior author Peter Noble of the University of Washington and Alabama State University told Seeker. “Different cell types have different life spans, generation times and resilience to extreme stress.”
In fact, some cells seem to fight to live after the organism has died.
“It is likely that some cells remain alive and are attempting to repair themselves, specifically stem cells,” Noble said.

For those poorly informed (educated) who insist that vaccines are just the same as catching the illness…. This is just one example of why that is not true.
Breakthrough for vaccine research: Mucosa forms special immunological memory
If a vaccine is to protect the intestines and other mucous membranes in the body, it also needs to be given through the mucosa, for example as a nasal spray or a liquid that is drunk. The mucosa forms a unique immunological antibody memory that does not occur if the vaccine is given by injection. This has been shown by a new study from Sahlgrenska Academy published in Nature Communications.
Immunological memory is the secret to human protection against various diseases and the success of vaccines. It allows our immune system to quickly recognize and neutralize threats. “The largest part of the immune system is in our mucosa. Even so, we understand less about how immunological memory protects us there than we do about protection in the rest of the body. Some have even suggested that a typical immune memory function does not exist in the mucosa,” says Mats Bemark, associate professor of immunology at Sahlgrenska Academy, University of Gothenburg.
After extensive work, the research team at Sahlgrenska Academy can now show that this assumption is completely wrong.
Mats Bemark et al. Limited clonal relatedness between gut IgA plasma cells and memory B cells after oral immunization, Nature Communications (2016). DOI: 10.1038/ncomms12698

Anti-microbial Peptides: Proteins that Pack a Punch
Antimicrobial resistance is a growing concern and it is currently estimated that approximately 2 million people are infected annually with serious infections that show antibiotic resistance to some degree. This contributes to the mortality of 23, 000 people with many more suffering severe complications as a direct result of antibiotic resistant infections. The economic burden on the US is thought to exceed $20 billion simply on health care bills alone, and a further $35 billion due to a societal loss in work based productivity (1).
The spread of antibiotic resistance is now widely believed to be a direct result of the anthropogenic release of antibiotics into the biosphere. We are now faced with the dilemma of how to treat these infections. In previous articles, I’ve talked largely about bacteriophages and how they are one possible solution to this complex problem. This article will introduce you to another class of antimicrobial agents, aptly called antimicrobial peptides (AMPs).
What are Antimicrobial Peptides?
Proteins are found ubiquitously throughout all cellular life and are like the mechanical parts of a car, helping your cells carry out a vast array of functions every single day. Peptides are small proteins that contain two or more amino acids joined by peptide bonds. Anyone who is familiar with biochemistry will be aware of the sheer diversity found amongst these versatile molecules. Needless to say, it should not be surprising that there are a large class of proteins involved in offensive cellular warfare. They are found widely in all domains of life and have evolved to give a cell a competitive advantage over its nastier neighbours.
Without getting too bogged down with the biochemistry, AMPs are characterised by their overall properties. AMPs that share common structural features will also have a similar function when targeting a cell. The diversity amongst these proteins can be seen in Figure 1, which shows some examples from the four classes of AMPs. The class I AMPs, the lantibiotics for example, all contain similar motifs which assign them a similar job. AMPs can range from anywhere between 6 to >59 amino acids, but are generally considered to be small proteins (2). They generally have a rather amphipathic nature and feature both positive and negative charges.
These peptides may have a number of rare (Figure 1), modified amino acids. The lanthionines are a class of AMP that contain lanthionine rings made from dehydrated serine and threonine residues connected by thioether cross-links. This happens after the protein leaves the ribosome and gives the protein some very unique properties which will be explained later in the article (3).
Figure 1. The four classes of AMPs, showing common examples in each class. Rare, modified amino acids are indicated by coloured circles with the three letter codes indicating the name of the residue. Thioether cross-links are indicated by an S coordinated by two black lines (3).
Implications for the Pharmaceutical industry
Our antibiotic pipeline is drying up (Figure 2), with few new drugs being approved by the Food and Drug Administration. Identifying novel antibiotics is a tedious process that requires a lot of time and effort from drug companies, which they are not willing to do. The reason for this boils down to economic reasons, as antibiotics are just not worth the investment. Unlike other drugs such as statins, antibiotics are only used for short periods of time by a patient. One course of treatment therefore doesn’t return a massive profit for the company. The second issue antibiotics face is that resistance to them occurs rapidly after they are put into circulation, so the company is not likely to get much use out of the drug. Therefore we need to find a new source for our antimicrobials. This is where the AMPs come in.
Currently, nearly 900 AMPs have been identified and characterised with many more undiscovered (2). They are an untapped source of drug discovery and they exhibit numerous benefits over their antibiotic cousins. As they are proteins, they have a genetic origin, which could provide an amenable platform for further development through random mutagenesis. This could produce a vast library of antimicrobial compounds (4,5), drastically improving our options for therapy.
Figure 2. Graph showing the steady decline in antibiotic development from 1980 to 2012 (1)
Nisin; not so nice if you’re a bacterial cell
AMPs were discovered in the 1930s although their use in the health industry has been fairly limited, resulting from the sheer difficulty and cost of manufacturing and purifying proteins on a large scale. The bacterially produced lantibiotics are by far the most well studied AMPs and have the most potential for the pharmaceutical industry. Nisin (E234) is the most well studied lantibiotic (Refer to Figure 1, Class I) and is produced by the bacterium Lactococcus lactis (6).
It shows broad spectrum activity on a large number of Gram-positive bacteria including other lactic acid bacteria, which has made it a coveted preservative in food processing. Currently it is added to cheeses, meats and beverages to extend shelf life and prevent the growth of spoilage organisms including spore forming bacteria such as Clostridium botulinum (6). The lantibiotics have also proven their capabilities for treating the clinically relevant pathogens methicillin-resistant Staphylococcus aureus and vancomycin-resistant Enterococci (7). They are also seen to have similar levels of activity as antibiotics and express low levels of toxicity to mammalian cells. Nisin exhibits poor oral availability making it more appealing as a topical agent or for intravenous application but there are also intentions to use it as a sterilising agent for catheters and medical equipment to help reduce the risk of infection (3).
So could lantibiotics like nisin be a good solution to antimicrobial resistance? Well more compelling evidence for nisin is that resistance has not been thoroughly documented. Nisin has been applied in sub-therapeutic concentrations in the food industry since the 1960s but still mostly retains its bactericidal ability. Resistance has been achieved artificially in the lab (discussed later in the article), but due to the mechanism of some lantibiotics including nisin, resistance is thought to be unlikely (6).
The mechanism behind nisin’s potency
Unlike animal cells, generally bacterial membranes have an overall negative charge and lack cholesterol (8). Nisin contains a high proportion of the positively charged (basic) amino acids lysine and arginine. These positive charges allow the protein to interact with the negative charges commonly associated with bacterial cell membranes (2). Nisin is good at aligning against Gram-positive bacterial membranes, where they multimerise to form short-lived pores (Figure 3). Hydrophobic regions help the protein to insert into the membrane and stabilise the pore (2), which allows the transport of ATP, ions and amino acids, eliminating the cellular membrane potential (9).
Nisin has a second trick up its sleeve. Its C-terminal, the portion of the protein containing the lanthionine ring motifs, allows it to latch onto the important membrane component lipid II (Figure 3). Lipid II is a precursor for peptidoglycan; the cell wall strengthening polymer found in both Gram-positive and Gram-negative bacteria. It is a common target for antibiotics including penicillin and vancomycin, which both target different stages of its synthesis. It helps to maintain the cell structure and prevents it from bursting under high osmotic pressure. When nisin binds to lipid II, it sequesters this molecule from the enzymes that catalyse its addition to growing peptidoglycan chains. Binding lipid II also helps to stabilise the transmembrane pores, further damaging the cell. As a result, not only is the cell wall weakened, but the cell loses its metabolic capabilities, through the loss of charged molecules.
The dual targeting system of nisin is thought to be the reason why resistance to nisin has not be well documented (10). The two processes are completely physiologically separate, and therefore to develop resistance, the bacteria would have to develop two unrelated mutations to counteract the effects of nisin.
Figure 3. Diagram showing the mechanism of several lantibiotics including nisin. AMPs are represented by lines made with clear circles. Phospholipids represented by green circles with tails. Lipid II is represented by orange hexagons (3).
What do we know about resistance towards nisin?
There are several proposed means by which an organism can be resistance to a toxin. Firstly, an organism may have innate immunity to a toxin simply because of its physiology. We see this largely in the Gram-negative bacteria towards nisin. The lipopolysaccharide (LPS) layer found on the outside of their cell wall provides protection against nisin and it has been shown that the oligosaccharides found within the core region of this structure greatly improve protection against nisin. It is believed that this is because metal ions are sequestered within this layer, adding additional positive charges to the site. Such charges would help to prevent nisin from aligning with the cell membrane (11). Removing these metal ions by sequestering them sensitises Gram-negative bacteria to nisin.
Emergent resistance is the type of resistance that should concern us the most, as it is the reason why we are now faced with the problem of antimicrobial resistance. It involves the acquisition of mutations or DNA that help confer tolerance to stress resulting from the action of a toxin (12). Although currently only produced in the laboratory, experiments carried out on the tolerance of clinically relevant bacteria towards nisin are crucial in highlighting the future of implementing an antimicrobial.
Resistance mechanisms have been documented in several bacteria including the causative agent of listeriosis, Listeria monocytogenes. Although not fully understood, changes in membrane composition have been attributed for the decreased susceptibility in resistant strains. In resistant strains, the bacterial membrane is composed of less negatively charged phospholipids. Similarly to sequestering metal ions near the membrane, this alters the overall net charge, helping to repel nisin.
The number of long chain fatty acids within its membrane is increased helping to reducing fluidity. This is believed to play a role in preventing nisin from inserting itself into the membrane. Studies show that nisin resistant strains were also less susceptible to cell wall acting components such as lysozyme and cell wall acting antibiotics. They did not identify the phenotypic change that gave additional protection, but this does indicate that a number of defence mechanisms are involved in defending cells against environmental stress from nisin (13).
Conclusion:
So could AMPs like nisin possibly serve as a replacement to our current armamentarium of antibiotics? AMPs are a largely untapped source of antimicrobials with many more still to be identified. AMPs may therefore serve as a new source of antimicrobials to help relieve the stress exerted on microorganisms by antibiotics. We have seen that nisin is an effective antimicrobial against a wide range of Gram-positive bacteria including spore forming bacteria. The dual-action of nisin challenges bacterial cells making it difficult for them to develop resistance. However, lab-based experiments have shown that it is possible to generate resistant strains showing the tenacity of bacteria to adapt to such potent environmental stresses. To learn from our previous mistakes with antibiotics, more responsible practices would need to be applied. Using combination therapy or rotating drug usage, as done with pesticides, could help further prevent resistance. Where they are likely to be applied in high concentration (in medical settings and agriculture), combination therapies should be used to further reduce the likelihood of resistance.
1. CDC. Antibiotic resistance threats. US Dep Healh Hum Serv. 2013;22–50.
2. Brogden KA. Antimicrobial peptides: pore formers or metabolic inhibitors in bacteria? Nat Rev Microbiol [Internet]. 2005;3(3):238–50. Available from: http://www.nature.com/doifinder/10.1038/nrmicro1098
3. Dischinger J, Basi Chipalu S, Bierbaum G. Lantibiotics: Promising candidates for future applications in health care. Int J Med Microbiol [Internet]. Elsevier GmbH.; 2014;304(1):51–62. Available from: http://dx.doi.org/10.1016/j.ijmm.2013.09.003
4. Field D, Begley M, O’Connor PM, Daly KM, Hugenholtz F, Cotter PD, et al. Bioengineered Nisin A Derivatives with Enhanced Activity against Both Gram Positive and Gram Negative Pathogens. PLoS One. 2012;7(10).
5. Hilpert K, Volkmer-Engert R, Walter T, Hancock REW. High-throughput generation of small antibacterial peptides with improved activity. Nat Biotechnol. 2005;23(8):1008–12.
6. van Heel AJ, Montalban-Lopez M, Kuipers OP. Evaluating the feasibility of lantibiotics as an alternative therapy against bacterial infections in humans. Expert Opin Drug Metab Toxicol. 2011;7(6):675–80.
7. Barbosa J, Caetano T, Mendo S. Class I and Class II Lanthipeptides Produced by Bacillus spp. J Nat Prod [Internet]. 2015;151008121848005. Available from: http://pubsdc3.acs.org/doi/10.1021/np500424y
8. Neumann A, Berends ETM, Nerlich A, Molhoek EM, Gallo RL, Meerloo T, et al. The antimicrobial peptide LL-37 facilitates the formation of neutrophil extracellular traps. Biochem J [Internet]. 2014 Nov 15 [cited 2014 Oct 28];464(1):3–11. Available from: http://www.ncbi.nlm.nih.gov/pubmed/25181554
9. Kordel M, Schuller F, Sahl HG. Interaction of the pore forming-peptide antibiotics Pep 5, nisin and subtilin with non-energized liposomes. FEBS Lett. 1989;244(1):99–102.
10. Islam MR, Nagao J, Zendo T, Sonomoto K. Antimicrobial mechanism of lantibiotics. Biochem Soc Trans [Internet]. 2012;40(6):1528–33. Available from: http://www.biochemsoctrans.org/bst/040/bst0401528.htm
11. Stevens K a., Sheldon BW, Klapes N a., Klaenhammer TR. Nisin treatment for inactivation of Salmonella species and other gram- negative bacteria. Appl Environ Microbiol. 1991;57(12):3613–5.
12. Kaur G, Malik RK, Mishra SK, Singh TP, Bhardwaj A, Singroha G, et al. Nisin and class IIa bacteriocin resistance among Listeria and other Foodborne pathogens and spoilage bacteria. Microb Drug Resist. 2011;17(2).
13. Crandall AD, Montville TJ. Nisin resistance in Listeria monocytogenes ATCC 700302 is a complex phenotype. Appl Environ Microbiol. 1998

Network Lost
If you believe the theory of six degrees of separation, we’re all connected to each other (and possibly to Kevin Bacon) by common friends and friend-of-friends. It might feel like a small world – in fact, these patterns crops up in all sorts of places. Small world networks connect distant brain cells, and help these lymph nodes (outlined in grey) fight infections. A network of fibroblastic reticular cells (FRCs, red) spreads out inside each node, producing chemicals (green) to support immune cells while they zip around the node gathering antigens – chemical information used to target bacteria. These mouse lymph nodes are treated with different doses of a toxin that destroys FRC networks. A high dose crumples the lymph node in the bottom right. Amazingly, many of these networks repair themselves, showing just how committed immune defences are to keeping their small worlds alive.
Written by John Ankers
Image from work by Mario Novkovic, Lucas Onder and Jovana Cupovic, and colleagues
Institute of Immunobiology, Kantonsspital St. Gallen, St. Gallen, Switzerland
Image originally published under a Creative Commons Licence (BY 4.0)
Published in PLOS Biology, July 2016
You can also follow BPoD on Twitter and Facebook


Meet the all-female team of coders that brought us Apollo 11.
In 1969, the world watched as Neil Armstrong marked his historic achievement with the words, “That’s one small step for man, one giant leap for mankind.” His now-famous transmission was heard around the globe thanks to NASA’s Deep Space Network, which made communication from outer space possible.
That network was built by a woman named Susan Finley. She was part of an all-female team of coders whose work was integral to the success of the Apollo 11 mission. Science writer Nathalia Holt brings us their stories in her book, Rise of the Rocket Girls: The Women Who Propelled Us from Missiles to the Moon to Mars.
Listen to their story here.
[Images via NASA]

Neurotransmitters are chemicals that help in transmitting signals across a synapse. Different neurotransmitters are associated with different functions. Knowledge about these helps us to treat various neurological conditions by either stimulating or inhibiting these production. #neurology #neuroscience #psychiatry #medicine #medstudynotes #medschool #mbbs #unimed #brain #nervoussystem #physiology #medblog #medblr #medstudent https://www.instagram.com/p/BrM4ocsBqJe/?utm_source=ig_tumblr_share&igshid=12tojib83c32d

I gotta split! Image of the Week - June 22, 2015
CIL:41466 - http://www.cellimagelibrary.org/images/41466
Description: Confocal image of a mitotic spindle in a dividing cell. The spindle is shown in yellow and the surrounding actin cytoskeleton is in blue. Sixth Prize, 2007 Olympus BioScapes Digital Imaging Competition.
Authors: Patricia Wadsworth and the 2007 Olympus Bioscapes Digital Imaging Competition®.
Licensing: Attribution Non-Commercial No Derivatives: This image is licensed under a Creative Commons Attribution, Non-Commercial, No Derivatives License
-
 dazehui liked this · 6 years ago
dazehui liked this · 6 years ago -
 b-duck liked this · 6 years ago
b-duck liked this · 6 years ago -
 bazeholdmybeer reblogged this · 6 years ago
bazeholdmybeer reblogged this · 6 years ago -
 noirecissist liked this · 6 years ago
noirecissist liked this · 6 years ago -
 queen-of-clubs-and-pans reblogged this · 6 years ago
queen-of-clubs-and-pans reblogged this · 6 years ago -
 queen-of-clubs-and-pans liked this · 6 years ago
queen-of-clubs-and-pans liked this · 6 years ago -
 themissdreamingstories liked this · 6 years ago
themissdreamingstories liked this · 6 years ago -
 dragonkinglover reblogged this · 6 years ago
dragonkinglover reblogged this · 6 years ago -
 dragonkinglover liked this · 6 years ago
dragonkinglover liked this · 6 years ago -
 ghoulish-activist-gay reblogged this · 6 years ago
ghoulish-activist-gay reblogged this · 6 years ago -
 ghoulish-activist-gay liked this · 6 years ago
ghoulish-activist-gay liked this · 6 years ago -
 aspensmonster liked this · 6 years ago
aspensmonster liked this · 6 years ago -
 darkstarbaby liked this · 7 years ago
darkstarbaby liked this · 7 years ago -
 kafanirabbi liked this · 7 years ago
kafanirabbi liked this · 7 years ago -
 forgetfultech liked this · 7 years ago
forgetfultech liked this · 7 years ago -
 di-mana liked this · 7 years ago
di-mana liked this · 7 years ago -
 bioengr reblogged this · 7 years ago
bioengr reblogged this · 7 years ago -
 cosmicvintage liked this · 7 years ago
cosmicvintage liked this · 7 years ago -
 sphereporcupine reblogged this · 8 years ago
sphereporcupine reblogged this · 8 years ago -
 sphereporcupine liked this · 8 years ago
sphereporcupine liked this · 8 years ago -
 applauseforsally liked this · 8 years ago
applauseforsally liked this · 8 years ago -
 concerned-relatives reblogged this · 8 years ago
concerned-relatives reblogged this · 8 years ago -
 disillusionedaydreamer liked this · 8 years ago
disillusionedaydreamer liked this · 8 years ago -
 astroanarchy reblogged this · 8 years ago
astroanarchy reblogged this · 8 years ago -
 wicketthicket liked this · 8 years ago
wicketthicket liked this · 8 years ago -
 cappucino-commie reblogged this · 8 years ago
cappucino-commie reblogged this · 8 years ago -
 spontaneous-rum reblogged this · 8 years ago
spontaneous-rum reblogged this · 8 years ago -
 494863 liked this · 8 years ago
494863 liked this · 8 years ago -
 mathsicalstudies-blog liked this · 8 years ago
mathsicalstudies-blog liked this · 8 years ago -
 kawuli reblogged this · 8 years ago
kawuli reblogged this · 8 years ago -
 brokentowels liked this · 8 years ago
brokentowels liked this · 8 years ago -
 thetravellingmooseys-blog liked this · 8 years ago
thetravellingmooseys-blog liked this · 8 years ago -
 weiyan520 liked this · 8 years ago
weiyan520 liked this · 8 years ago -
 knead-weed reblogged this · 8 years ago
knead-weed reblogged this · 8 years ago -
 watsdaughter liked this · 8 years ago
watsdaughter liked this · 8 years ago -
 circularpi liked this · 8 years ago
circularpi liked this · 8 years ago -
 blissburrito liked this · 8 years ago
blissburrito liked this · 8 years ago -
 higgs-boson23 liked this · 8 years ago
higgs-boson23 liked this · 8 years ago -
 saturnusminor liked this · 8 years ago
saturnusminor liked this · 8 years ago -
 somthingclever-blog1 liked this · 8 years ago
somthingclever-blog1 liked this · 8 years ago -
 mosieart reblogged this · 8 years ago
mosieart reblogged this · 8 years ago -
 tescobar3 liked this · 8 years ago
tescobar3 liked this · 8 years ago -
 austinswart liked this · 8 years ago
austinswart liked this · 8 years ago -
 delicious-gluten reblogged this · 8 years ago
delicious-gluten reblogged this · 8 years ago
A pharmacist and a little science sideblog. "Knowledge belongs to humanity, and is the torch which illuminates the world." - Louis Pasteur
215 posts